INTRODUCTION
Evolution of methicillin-resistant Staphylococcus aureus (MRSA) against beta-lactam antibiotics is a challenging issue for clinicians in the management of infections.[1] S. aureus is one of the major pathogens causes wide range of minor and major infections leading to morbidity and mortality. Penicillin invention reduced the mortality rates of infectious diseases including S. aureus infection.[2] Widespread use of antibiotics results in the evolution of more resistant strains. An organism can become resistant to a particular antibiotic through repeated exposure. The resistance is a result of genetic alterations or transfer of resistant gene.[3] S. aureus acquired resistance to beta-lactam antibiotics by harboring a gene called mecA. This restricts the use of beta-lactam antibiotics against MRSA infections.[4]
mecA is the gene responsible for resistance against beta-lactam antibiotics in S. aureus. mecA encodes a unique (altered) penicillin-binding protein (PBP) called PBP2 or MRSA-PBP, which executes low affinity to almost all beta-lactam antibiotics.[5]
A 21-60 kb DNA segment called chromosome cassette mec CCMec harbors the mecA gene along with other genes encoding resistance to non-beta-lactam antibiotics. mecA is a 2010 bp gene flanking from 47917 to 49926 in the genomic DNA of MRSA, which ensures resistance to beta-lactams. The gene product is a 78kDa protein PBP2 which is a modified form of native PBP. Methicillin-sensitive S. aureus (MSSA) encodes the native PBP and beta-lactams bind and disrupt the peptidoglycan layer synthesis. In MRSA, beta-lactam antibiotics cannot bind to the modified PBP (PBP2). Hence cell wall synthesis continues even in the presence of antibiotic.[4,6] Through horizontal gene transfer, mecA could be transferred from MRSA to MSSA by bacterial conjucation. This mechanism increases the prevalence of beta-lactam resistant strains among nosocomial infections which results in the failure of antibiotic therapy.[7,8] Identifying methicillin resistance in S. aureus infections is essential to choose right antibiotics for treatment.
Exposure of MRSA to beta-lactam antibiotics results in the production of PBPs excessively. It has been found that the induction of mecA and penicillinase production occurs simultaneously in the presence of beta-lactam antibiotics. This explains the fact that mechanism behind the stimulation and overexpression of PBP and penicillinase would be the same.[9]
MATERIALS AND METHODS
Sampling
94 strains of S. aureus isolated from minor wounds and postoperative wounds of patients, attending a rural tertiary care hospital were included in the study.
Wound swabs were collected and 5% blood agar and MacConkey quadrant streaking was for the isolation of bacteria following standard operating procedures. S. aureus was identified by slide and tube coagulase tests.
Antibiotic Sensitivity Assay
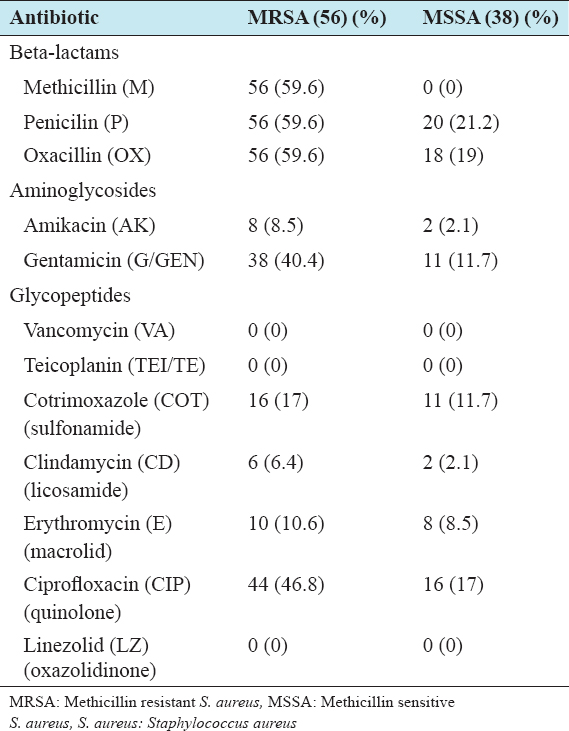
All isolates were subjected to antibiotic sensitivity test by modified Kirby-Bauer technique based on NCCLS regulations. The antibiotic panel includes, methicillin (M), penicillin (P), oxacillin (OX), amikacin (AK), Co-trimoxazole (COT), gentamicin (G/GEN), vancomycin (VA), clindamycin (CD), erythromycin (E), teicoplanin (TEI/TE), ciprofloxacin (CIP), cefazolin (CZ), cefoxitin (CX), and linezolid (LZ) and the isolates were observed as sensitive or resistant based on the measurement of zone of inhibition (Table 1).
Table 1: Antibiotic resistance pattern of S. aureus isolates
DNA Isolation and mecA Gene Amplification
DNA was isolated by rapid heat and cool method. A loopful of bacterial culture was transferred to 100 µl of 50 mM NaoH and incubated at 95°C for 5 min. The sample was immediately transferred to 4°C. It was followed by the addition of 20 µl of 1Mtris HCL and centrifugation at 8000RPM for 3 min. The supernatant containing DNA was transferred to a fresh tube.
RESULTS
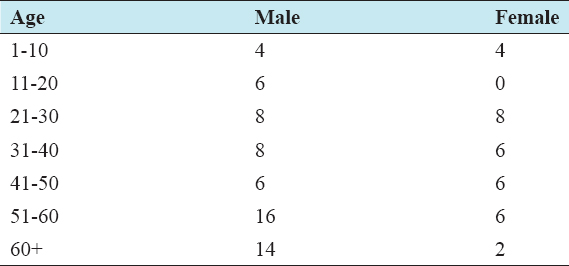
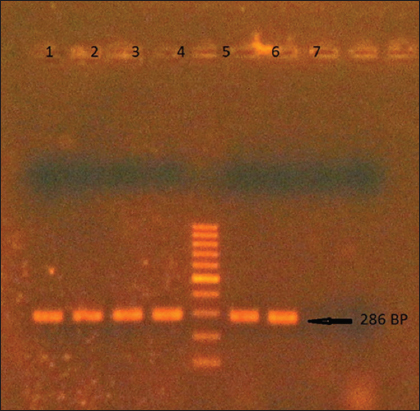
S. aureus infection was predominant in males (62) than females (32). The study population was aged from a minimum of 1 to a maximum of 74 years with an average of 39 years. More number of isolates were obtained from older age groups of 50+ years (Table 2). Polymerase chain reaction (PCR) amplified a 286 bp sequence of the mecA gene in 56 of the 94 isolates (59.6%), which was confirmed in 1% agarose gel electrophoresis (Figure 1). All MRSA identified by PCR were resistant to methicillin with varying inhibitory concentrations by disc diffusion.
Table 2: Age and sex distribution of Staphylococcal infection
Figure 1: Amplified product of mecA gene in 1% agarose gel. Lane 5: 100bp DNA marker
Amplification of mecA gene was done using primers described previously, F: 5’-TGCTATCCACCCTCAAACAGG-3’, R: 5’-AACGTTGTAACCACCCCAAGA-3’ (Kondo et al., 2007). The 286 bp sequence was amplified with 500 µm of each forward and reverse primer, 5 µl of template DNA were used in a 50 µl of 1X master mix (Takara- EmraldAmp). Amplification was done in BIO-RAD T100 thermal cycler with 35 cycles of initial denaturation at 93°C for 5 min, cycle denaturation at 93°C for 1 min, annealing at 55°C for 1 min, cycle extension at 72°C for 1 min, final extension at 72°C for 5 min and hold at 4°C for 5 min. The resulting product was viewed in 1% agarose gel with 100bp DNA marker.
The resistance rate of both MRSA and MSSA were more against penicillin and oxacillin followed by ciprofloxacin and gentamicin. Linezolid, vancomycin and teicoplanin were equally effective against both MRSA and MSSA. None of the isolates were resistant against linezolid, vancomycin and teicoplanin (Table 1).
DISCUSSION
The prevalence of hospital-acquired MRSA is more than the previous reports available in the literature across the country.[10,11] This shows that the prevalence rate is increasing gradually over time. Global prevalence rates vary among countries from a minimum of <1% to higher rates of more than 50%.[12] The infection rate of our study was found to be more in males than in females like the other community-based studies.[6,13,14] This study indicates MRSA is more common in wound infections and open wounds are more susceptible to acquire hospital based transmission of MRSA. Thereby supports the report of Kini et al. 2013, who have observed the prevalence of MRSA in joint wounds of children in India.[15]
The PCR protocol amplified the mecA gene in all methicillin-resistant strains identified through disc diffusion analysis. Both PCR and disc diffusion tests are equally efficient, and PCR is more rapid than the culture protocols, by which results can be obtained within few hours. The antibiotic resistance pattern shows that all mecA positive strains are phenotypically resistant to penicillin and oxacillin. Some mecA negative isolates were also observed to be resistant to penicillin and oxacillin. This could be explained by the fact that production of beta-lactamase but not the novel PBP, would contribute to the resistance in these strains.[16]
Glycopeptides; both vancomycin and teicoplanin exhibited 100% activity against MRSA. Vancomycin has been the drug of choice for MRSA infections, but vancomycin-resistant S. aures (VRSA) also emerged as a new challenge in infection management.[17] However, we have not found any VRSA strains among the isolates included in the study.
S. aureus more quickly develops resistance to most of the commonly used antibiotics. This creates a necessity to use more recent antibiotics like linezolid. Linezolid-resistant S. aureus (LRSA) has been found to be <1%. However higher prevalence of around 40% LRSA was reported from North America and Europe. The existance of resistance to linezolid in LRSA is a mutation in the 23srRNA gene.[18,19]
CONCLUSION
Prevalence of hospital-associated MRSA is increasing drastically in the past few decades. More precautions and aseptic procedures should be followed in hospitals to minimize the spread of MRSA. Both phenotypic and genotypic determination of MRSA is recommended, and mecA PCR is found to be a rapid and reliable method. The presence of mecA gene in all MRSA strains indicates mecA is responsible for methicillin resistance. According to the present study, MRSA was found to be resistant to all beta-lactams. Vancomycin, teicoplanin and linezolid were 100% effective against all the hospital isolated strains and recommended as drugs of choice for MRSA.